UW Interactive Data Lab
papers

invis: Exploring High-Dimensional RNA Sequences from In Vitro Selection
Çağatay Demiralp, Eric Hayden, Jeff Hammerbacher, Jeffrey Heer.
Proc. IEEE Biological Data Visualization (BioVis), 2013
Çağatay Demiralp, Eric Hayden, Jeff Hammerbacher, Jeffrey Heer
Proc. IEEE Biological Data Visualization (BioVis), 2013
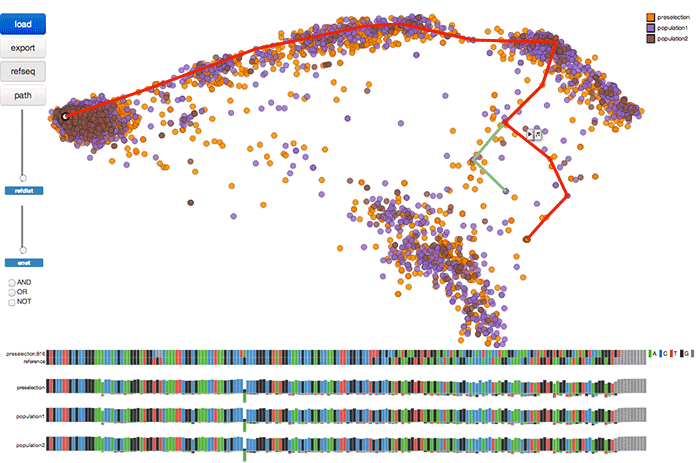
Abstract
In vitro selection and evolution is a powerful method for discovering RNA molecules based on their binding and catalysis properties. It has important applications to the study of genetic variation and molecular evolution. However, the resulting RNA sequences form a large, high-dimensional space and biologists lack adequate tools to explore and interpret these sequences. We present invis, the first visual analysis tool to facilitate exploration of in vitro selection sequence spaces. invis introduces a novel configuration of coordinated views that enables simultaneous inspection of global projections of sequence data alongside local regions of selected dimensions and sequence clusters. It allows scientists to isolate related sequences for further data analysis, compare sequence populations over varying conditions, filter sequences based on their similarities, and visualize likely pathways of genetic evolution. User feedback indicates that invis enables effective exploration of in vitro RNA selection sequences.
BibTeX
@inproceedings{2013-invis,
title = {invis: Exploring High-Dimensional RNA Sequences from In Vitro Selection},
author = {Demiralp, \c{C}a\u{g}atay AND Hayden, Eric AND Hammerbacher, Jeff AND Heer, Jeffrey},
booktitle = {Proc. IEEE Biological Data Visualization (BioVis)},
year = {2013},
url = {https://idl.uw.edu/papers/invis},
doi = {10.1109/BioVis.2013.6664340}
}
